4 Epigenomes Landscapes of CCOC models
4.1 Loading Data
4.2 Descriptive Summary
4.2.1 Descriptive Summary of H3K27ac Peaks
peak.summary.list <- lapply(gr.list,get.genomeCoverage)
peak.summary.df <- data.frame(do.call(rbind,peak.summary.list))
peak.summary.df$widthsd <- lapply(peak.summary.df$width,sd)
#apply element-wise divsion of vector scores and vector widthpeak by row
peak.summary.df$singal.per.width <-apply(peak.summary.df[c("scores","widthPeak")], 1,function(x){unlist(x[1],use.names = FALSE)/unlist(x[2],use.names = FALSE)} )
peak.summary.df <- cbind(peak.summary.df, t(sapply(peak.summary.df$widthPeak,summary)))
peak.summary.df$target <- rownames(peak.summary.df) %>% str_replace(.,"[2-3]D[-,_]","")
peak.summary.df$condition <- CCOC_2D_3D_pooled$condition
peak.summary.df <- peak.summary.df %>% mutate_at(c("condition","numPeak"),unlist)
kable(peak.summary.df %>% dplyr::select(-c(scores,widthPeak,singal.per.width,target,condition)))%>% add_header_above(header = c(" " = 3, "Width Summary"= 7)) %>% kable_styling("striped") %>%scroll_box(width = "100%")| numPeak | percentCoverage | widthsd | Min. | 1st Qu. | Median | Mean | 3rd Qu. | Max. | |
|---|---|---|---|---|---|---|---|---|---|
| 3D-HAC2 | 36315 | 1.014851 | 730.148 | 285 | 409 | 625.0 | 847.0485 | 1030.50 | 21877 |
| 3D-JHOC9 | 26182 | 0.5893284 | 633.499 | 200 | 315 | 493.0 | 682.2548 | 817.00 | 14561 |
| 3D_KOC-7 | 32459 | 2.432168 | 2191.885 | 415 | 949 | 1774.0 | 2271.1738 | 2785.00 | 44541 |
| 3D_OV207 | 50210 | 1.440366 | 991.7094 | 168 | 308 | 546.0 | 869.5099 | 1060.00 | 26496 |
| 3D_OVISE | 36591 | 1.39044 | 1216.069 | 185 | 368 | 752.0 | 1151.7810 | 1551.50 | 25978 |
| 3D_SMOV2 | 37888 | 1.766143 | 1634.804 | 230 | 451 | 918.0 | 1412.9151 | 1783.00 | 41833 |
| 3D-C5X | 27040 | 0.8671941 | 1017.543 | 212 | 379 | 662.0 | 972.0792 | 1219.00 | 31482 |
| 3D-HCH1 | 37424 | 1.379521 | 1359.268 | 188 | 371 | 683.0 | 1117.3010 | 1377.00 | 33858 |
| 2D_C5X | 35938 | 1.54898 | 1546.566 | 190 | 404 | 846.0 | 1306.4231 | 1686.00 | 68650 |
| 2D_ES2 | 27788 | 1.512809 | 1464.576 | 318 | 716 | 1319.5 | 1650.1329 | 2042.00 | 26983 |
| 2D_HAC2 | 34440 | 1.5771 | 1548.123 | 190 | 424 | 944.0 | 1387.9955 | 1826.00 | 31942 |
| 2D_HCH1 | 45473 | 1.901269 | 1465.685 | 195 | 397 | 788.0 | 1267.3075 | 1652.00 | 41766 |
| 2D_JHOC5 | 34157 | 1.499355 | 1288.799 | 192 | 443 | 975.0 | 1330.5056 | 1783.00 | 24046 |
| 2D_JHOC9 | 42100 | 1.579591 | 1283.515 | 190 | 380 | 705.0 | 1137.2463 | 1455.25 | 44197 |
| 2D_KOC-7 | 28050 | 1.6566 | 1596.533 | 262 | 733 | 1499.0 | 1790.0980 | 2278.00 | 42949 |
| 2D_OV207 | 39529 | 2.008683 | 1636.637 | 218 | 507 | 1128.0 | 1540.2371 | 1963.00 | 30344 |
| 2D_OVAS | 32562 | 1.546362 | 1405.513 | 228 | 498 | 1047.0 | 1439.4354 | 1891.00 | 36267 |
| 2D_OVISE | 27756 | 1.251867 | 1267.911 | 240 | 530 | 1055.0 | 1367.0781 | 1774.00 | 27178 |
| 2D_OVMANA | 37724 | 1.614853 | 1391.516 | 200 | 417 | 866.0 | 1297.5000 | 1723.25 | 31266 |
| 2D_OVSAYO | 39968 | 1.798394 | 1412.501 | 218 | 456 | 935.0 | 1363.8432 | 1786.00 | 33122 |
| 2D_OVTOKO | 45950 | 1.979838 | 1392.87 | 188 | 405 | 844.0 | 1305.9789 | 1742.00 | 28207 |
| 2D_RMGII | 46439 | 2.180606 | 1649.837 | 192 | 434 | 899.0 | 1423.2669 | 1862.00 | 43923 |
| 2D_SMOV2 | 39757 | 1.984135 | 1680.421 | 230 | 483 | 982.0 | 1512.6890 | 1946.00 | 44701 |
| 2D_TOV21G | 33000 | 1.805953 | 1976.66 | 242 | 531 | 1178.5 | 1658.7637 | 2096.00 | 91595 |
select_col <- c("target","widthPeak","singal.per.width","condition","Median","scores")
peak.volin.df <- peak.summary.df[select_col] %>% unnest(cols = c(widthPeak, singal.per.width, scores))
xlabs <- paste(unique(peak.volin.df$condition),"\n(N=",table(peak.summary.df$condition)%>% sort,")",sep="")
ggplot(peak.summary.df, aes(x=condition, y=numPeak, fill= condition))+ scale_x_discrete(labels=xlabs)+geom_boxplot(width=0.5,position = position_dodge(0.8))+ geom_dotplot(binaxis = "y", stackdir='center', dotsize = 1,position = position_dodge(0.8))+ labs(y="# of Peak") + theme_classic()## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.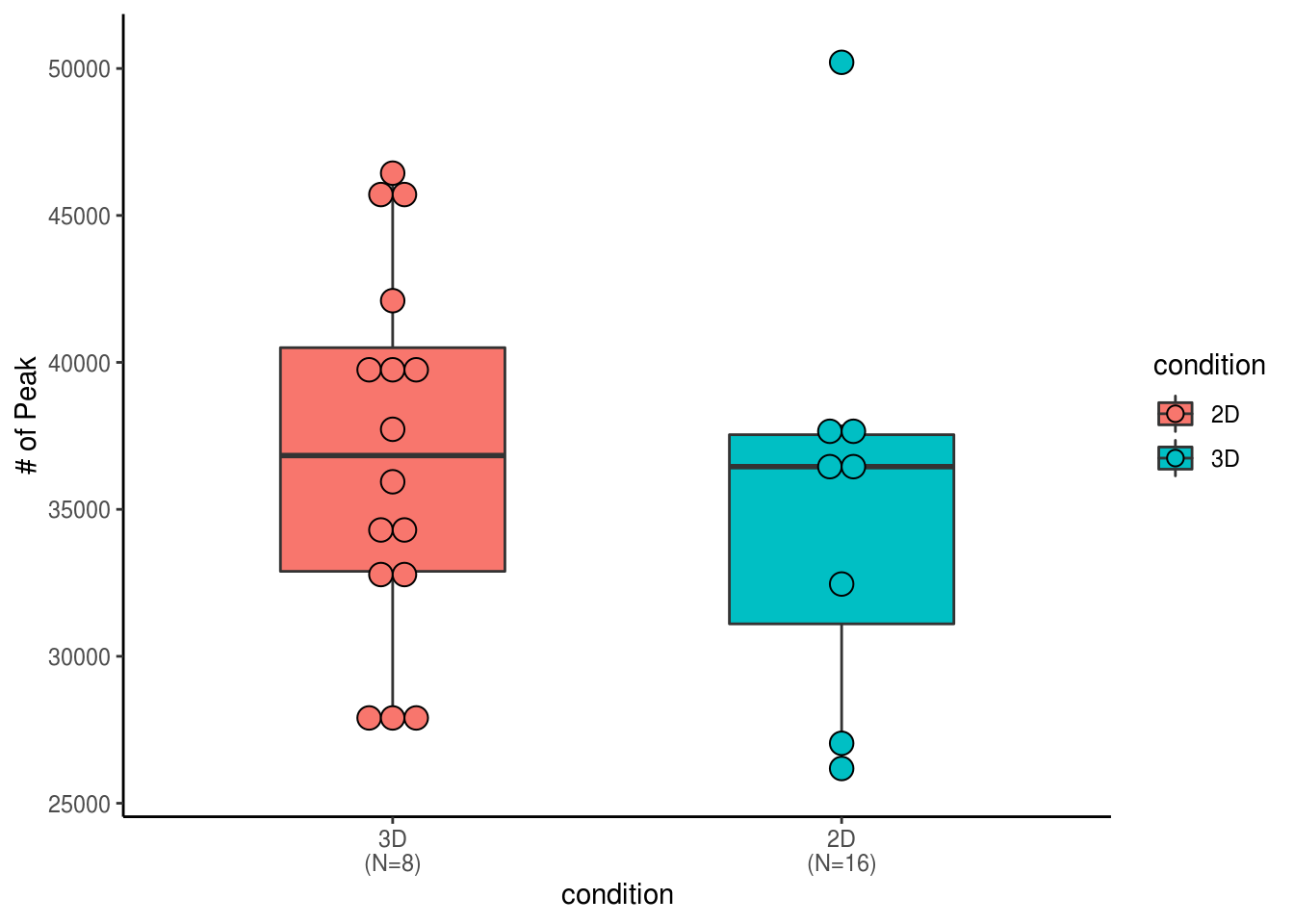
## notch went outside hinges. Try setting notch=FALSE.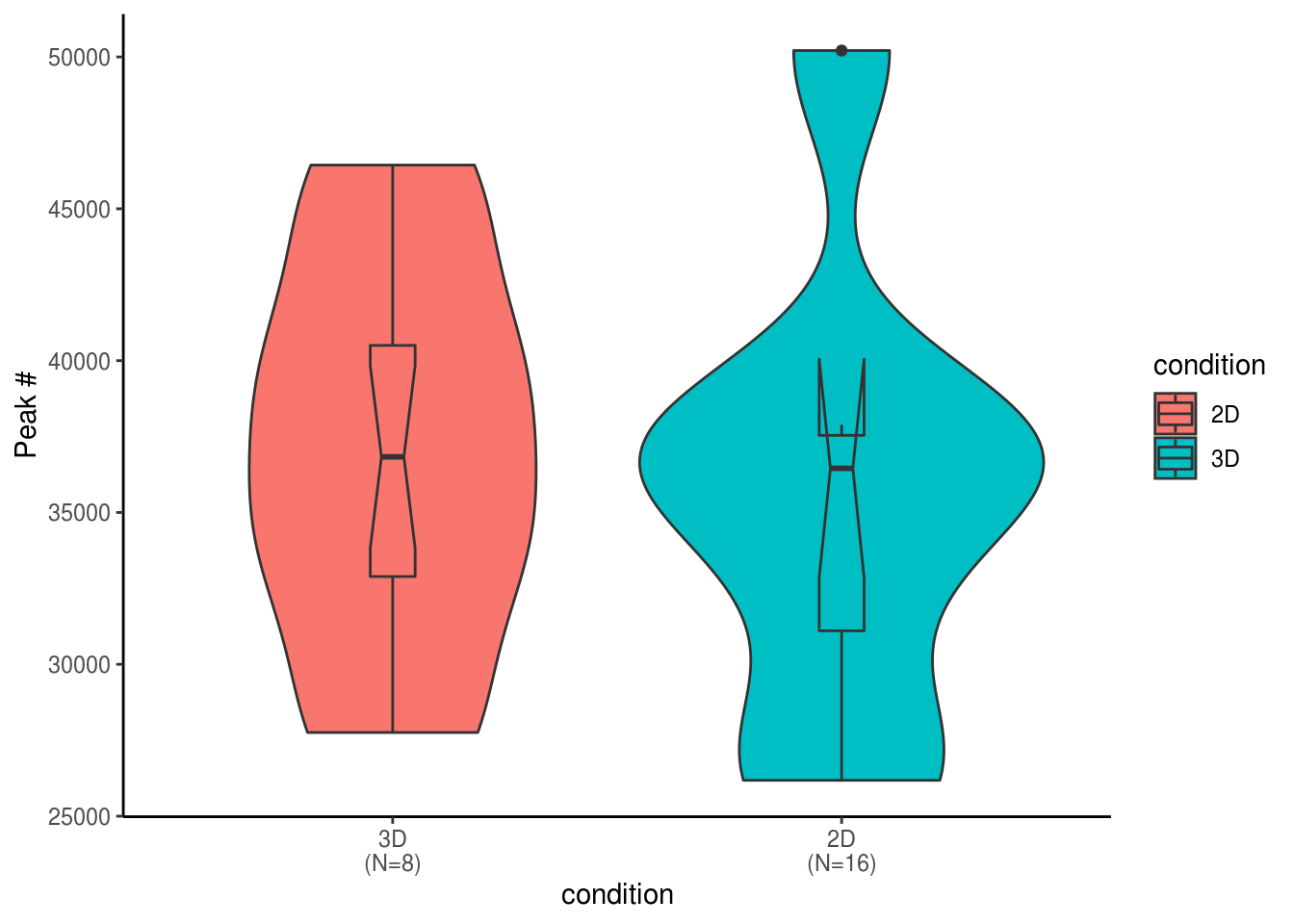
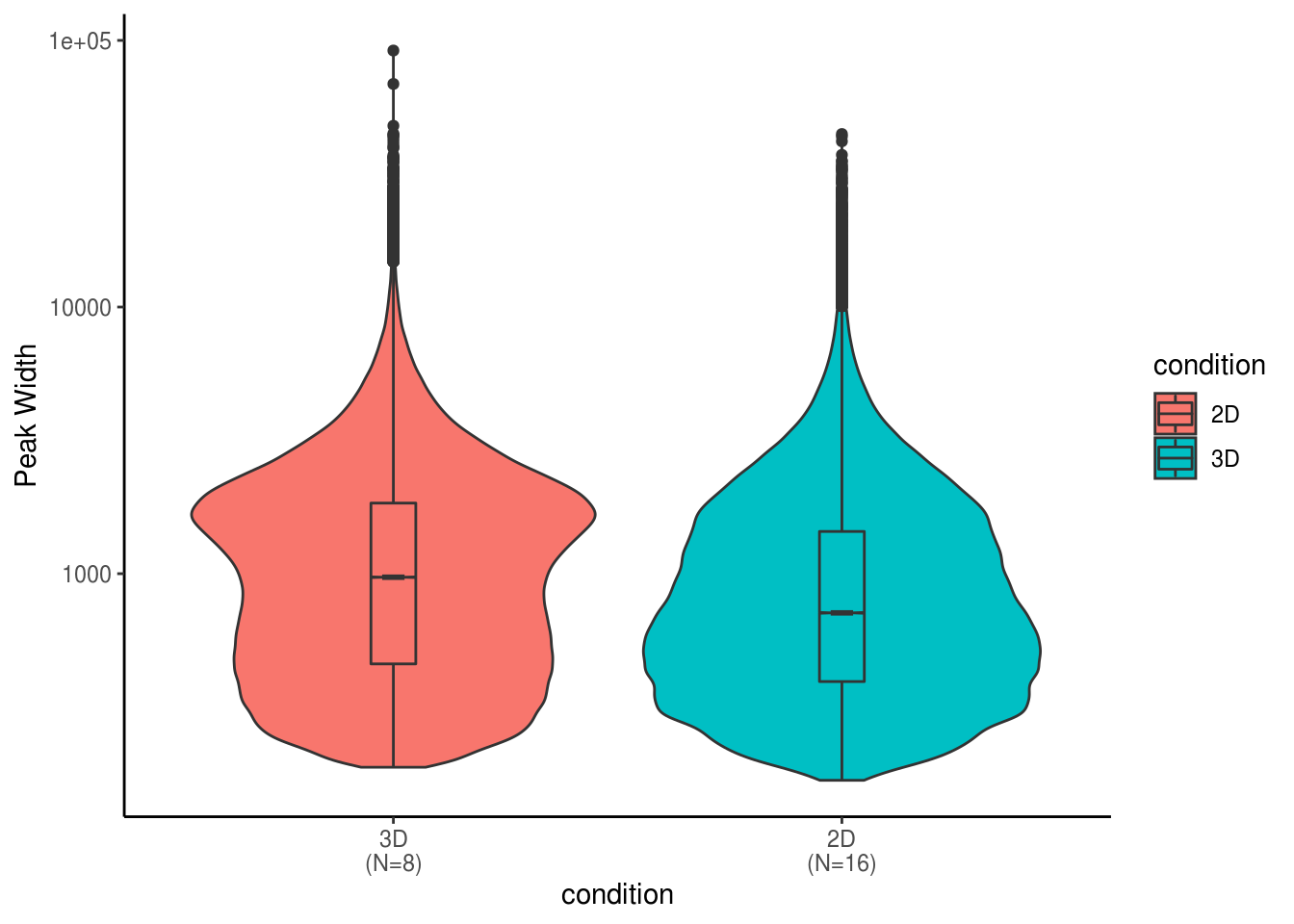
4.2.2 Descriptive Summary of Super Ehancer Peaks
dir="data/ChIP-seq/rose2"
AE.files <- list.files(path = dir, pattern = "*_AllEnhancers.table.txt",recursive = TRUE , full.names=TRUE )
all.table <- cbind(dir=dirname(AE.files),all=AE.files)
SE.gene.file <- list.files(path = dir, pattern = "*_SuperEnhancers_ENHANCER_TO_GENE.txt",recursive = TRUE, full.names=TRUE)
se.table <- cbind(dir=dirname(SE.gene.file), se_gene=SE.gene.file)
se.table <- merge(se.table,all.table,by="dir")
se.table$sample <- se.table$dir %>% str_replace("/rep","_rep") %>% str_replace("_R[1,2]_00[0-9]","") %>% str_replace("-","_") %>% basename()
se.table$condition <- se.table$sample %>% str_extract("^[2-3]D")
se.table <- se.table %>% filter(str_detect(se.table$sample,str_c(CCOC_2D_3D_pooled$sample, collapse ="|"))) %>% filter( sample %notin% c("3D-HAC2-B-Ac_rep1", "3D-JHOC9-B-Ac_rep2"))se.gr <- sapply(se.table$se_gene, read.rose2.output,select.meta.col=c("isSuper","OVERLAP_GENES","PROXIMAL_GENES") )
peak.summary.list <- lapply(se.gr,get.genomeCoverage)
peak.summary.df <- data.frame(do.call(rbind,peak.summary.list))
peak.summary.df$widthsd <- lapply(peak.summary.df$width,sd)
#apply element-wise divsion of vector scores and vector widthpeak by row
#peak.summary.df$singal.per.width <-apply(peak.summary.df[c("scores","widthPeak")], 1,function(x){unlist(x[1],use.names = FALSE)/unlist(x[2],use.names = FALSE)} )
peak.summary.df <- cbind(peak.summary.df, t(sapply(peak.summary.df$widthPeak,summary)))
peak.summary.df$target <- rownames(peak.summary.df) %>% str_replace(.,"[2-3]D[-,_]","")
peak.summary.df$condition <- se.table$condition
rownames(peak.summary.df) <- se.table$sample
kable(peak.summary.df %>% dplyr::select(-c(scores,widthPeak,target,condition)))%>% add_header_above(header = c(" " = 3, "Width Summary"= 7)) %>% kable_styling("striped") %>%scroll_box(width = "100%")| numPeak | percentCoverage | widthsd | Min. | 1st Qu. | Median | Mean | 3rd Qu. | Max. | |
|---|---|---|---|---|---|---|---|---|---|
| 2D_C5X-A_rep1 | 521 | 0.5119433 | 18565.43 | 3757 | 15569.00 | 26111.0 | 29783.53 | 39958.00 | 108224 |
| 2D_C5X-A_rep2 | 565 | 0.5696623 | 20740.11 | 3556 | 15157.00 | 26217.0 | 30560.54 | 40323.00 | 174900 |
| 2D_ES2-A_rep1 | 390 | 0.3499588 | 17544.95 | 3534 | 14459.75 | 23555.0 | 27198.46 | 35448.75 | 107312 |
| 2D_ES2-A_rep2 | 410 | 0.377693 | 18304.07 | 3083 | 14733.75 | 23400.0 | 27922.03 | 37709.00 | 107591 |
| 2D_HAC2-A_rep1 | 668 | 0.4848926 | 16587.81 | 2752 | 9222.00 | 18638.5 | 22001.95 | 29631.25 | 109913 |
| 2D_HAC2-A_rep2 | 751 | 0.5457887 | 17843.16 | 2515 | 8510.50 | 17103.0 | 22028.08 | 28994.00 | 120111 |
| 2D_HCH1-A_rep1 | 1064 | 0.8061596 | 17349.01 | 1633 | 9597.50 | 19032.5 | 22965.26 | 30798.75 | 152009 |
| 2D_HCH1-A_rep2 | 1082 | 0.8736538 | 18354.24 | 2333 | 10660.75 | 20402.5 | 24473.95 | 32972.75 | 153950 |
| 2D_JHOC5-A_rep1 | 456 | 0.4008345 | 19314.41 | 3613 | 13062.75 | 22367.0 | 26643.56 | 34618.00 | 138949 |
| 2D_JHOC5-A_rep2 | 435 | 0.4006083 | 18905.28 | 3906 | 14471.00 | 24267.0 | 27914.04 | 36174.00 | 119538 |
| 2D_JHOC9-A_rep1 | 752 | 0.6029846 | 16508 | 3217 | 11862.75 | 20675.5 | 24304.15 | 33036.50 | 103012 |
| 2D_JHOC9-A_rep2 | 768 | 0.5927454 | 16022.95 | 3180 | 11389.50 | 19661.0 | 23393.70 | 31899.00 | 96106 |
| 2D_KOC-7c-A_rep1 | 702 | 0.4901483 | 15289.26 | 3159 | 9755.00 | 17601.5 | 21163.25 | 28316.75 | 103395 |
| 2D_KOC-7c-A_rep2 | 779 | 0.5688513 | 16059.52 | 3028 | 10602.00 | 18500.0 | 22133.66 | 28890.00 | 106531 |
| 2D_OV207-A_rep1 | 387 | 0.4684759 | 22306.2 | 4332 | 20240.50 | 31670.0 | 36691.74 | 49049.50 | 156318 |
| 2D_OV207-A_rep2 | 420 | 0.5049983 | 21992.37 | 4154 | 20100.50 | 31410.5 | 36444.55 | 48506.25 | 135536 |
| 2D_OVAS-A_rep1 | 704 | 0.598128 | 19269.76 | 2035 | 11310.75 | 21653.0 | 25752.15 | 35771.50 | 146237 |
| 2D_OVAS-A_rep2 | 652 | 0.5679318 | 19773.7 | 1951 | 11710.75 | 22206.0 | 26402.23 | 36384.00 | 151425 |
| 2D_OVISE-A_rep1 | 304 | 0.2321447 | 17404.65 | 2679 | 9467.25 | 19514.0 | 23146.07 | 32350.25 | 108359 |
| 2D_OVISE-A_rep2 | 325 | 0.2716723 | 18694.39 | 3144 | 10603.00 | 21365.0 | 25336.93 | 35308.00 | 110762 |
| 2D_OVMANA-A_rep1 | 896 | 0.6942538 | 17307.77 | 1795 | 10768.50 | 19086.5 | 23485.63 | 31545.00 | 126026 |
| 2D_OVMANA-A_rep2 | 820 | 0.635191 | 17097.79 | 1730 | 11007.25 | 18995.0 | 23479.16 | 30770.50 | 107370 |
| 2D_OVSAYO-A_rep1 | 683 | 0.6029002 | 18840.11 | 3112 | 12163.00 | 22791.0 | 26755.72 | 37075.50 | 138702 |
| 2D_OVSAYO-A_rep2 | 917 | 0.7466912 | 19185.13 | 2661 | 10394.00 | 20138.0 | 24681.05 | 33714.00 | 170072 |
| 2D_OVTOKO-A_rep1 | 807 | 0.7221738 | 18949.59 | 3598 | 13412.50 | 22970.0 | 27124.40 | 36636.50 | 173712 |
| 2D_OVTOKO-A_rep2 | 523 | 0.6499654 | 23446.06 | 4587 | 22014.00 | 32997.0 | 37668.69 | 47481.50 | 175160 |
| 2D_RMGII-A_rep1 | 563 | 0.6376633 | 21335.14 | 3813 | 18576.00 | 30242.0 | 34330.10 | 44751.00 | 136243 |
| 2D_RMGII-A_rep2 | 490 | 0.5758231 | 21509.06 | 3663 | 19582.50 | 31580.5 | 35619.27 | 46039.75 | 136470 |
| 2D_SMOV2-A_rep1 | 615 | 0.5633793 | 17450.2 | 3814 | 14485.00 | 23985.0 | 27766.29 | 37245.50 | 103719 |
| 2D_SMOV2-A_rep2 | 599 | 0.5498222 | 18009.13 | 3910 | 14603.00 | 23954.0 | 27821.94 | 36817.50 | 122125 |
| 2D_TOV21G-A_rep1 | 315 | 0.5014834 | 34021.29 | 4062 | 22744.50 | 41689.0 | 48254.52 | 66369.50 | 288049 |
| 2D_TOV21G-A_rep2 | 309 | 0.4876389 | 33720.33 | 4940 | 23917.00 | 41503.0 | 47833.47 | 63885.00 | 289296 |
| 3D_KOC-7C-A_rep1 | 616 | 0.5802729 | 18514.43 | 4269 | 14931.50 | 25444.5 | 28552.46 | 37722.25 | 153761 |
| 3D_KOC-7C-A_rep2 | 723 | 0.857726 | 24699.55 | 4478 | 17682.00 | 30106.0 | 35958.56 | 47761.00 | 181481 |
| 3D_OV207-A_rep1 | 277 | 0.3917063 | 25154.18 | 3638 | 25500.00 | 38303.0 | 42862.04 | 52458.00 | 185725 |
| 3D_OV207-A_rep2 | 403 | 0.3844735 | 18398.1 | 3657 | 16648.00 | 24900.0 | 28917.01 | 36505.50 | 154766 |
| 3D_OVISE-A_rep1 | 575 | 0.5493934 | 22011.9 | 3529 | 11879.50 | 23856.0 | 28960.60 | 40605.50 | 185088 |
| 3D_OVISE-A_rep2 | 550 | 0.4348488 | 17754.91 | 3508 | 10296.25 | 20138.5 | 23964.46 | 32015.50 | 118553 |
| 3D_SMOV2-A_rep1 | 563 | 0.6481212 | 21923.11 | 4211 | 18525.00 | 30399.0 | 34893.12 | 45388.50 | 131706 |
| 3D_SMOV2-A_rep2 | 710 | 0.7388403 | 21746.27 | 3184 | 15472.75 | 26197.0 | 31541.64 | 41969.75 | 144659 |
| 3D-C5X-B-Ac_rep1 | 343 | 0.3424175 | 16394.4 | 6428 | 19250.00 | 27260.0 | 30258.95 | 37272.00 | 126290 |
| 3D-C5X-B-Ac_rep2 | 380 | 0.2360343 | 11944.55 | 3175 | 10472.25 | 16643.5 | 18827.11 | 23923.50 | 78719 |
| 3D-HAC2-B-Ac_rep2 | 735 | 0.8258416 | 25638.32 | 1765 | 18016.00 | 29220.0 | 34056.61 | 43685.00 | 225683 |
| 3D-HCH1-B-Ac_rep1 | 570 | 0.529702 | 17090.01 | 3648 | 17030.00 | 24722.0 | 28167.53 | 34956.75 | 153872 |
| 3D-HCH1-B-Ac_rep2 | 834 | 0.7880745 | 19031.3 | 3721 | 14906.25 | 24137.0 | 28641.33 | 37442.00 | 125041 |
| 3D-JHOC9-B-Ac_rep1 | 358 | 0.2719044 | 13240.78 | 4503 | 14175.75 | 20032.5 | 23021.05 | 28950.75 | 92878 |
select_col <- c("target","widthPeak","condition","Median")
peak.volin.df <- peak.summary.df[select_col] %>% unnest(cols = c(widthPeak))
xlabs <- paste(unique(peak.volin.df$condition),"\n(N=",table(peak.summary.df$condition)%>% sort,")",sep="")
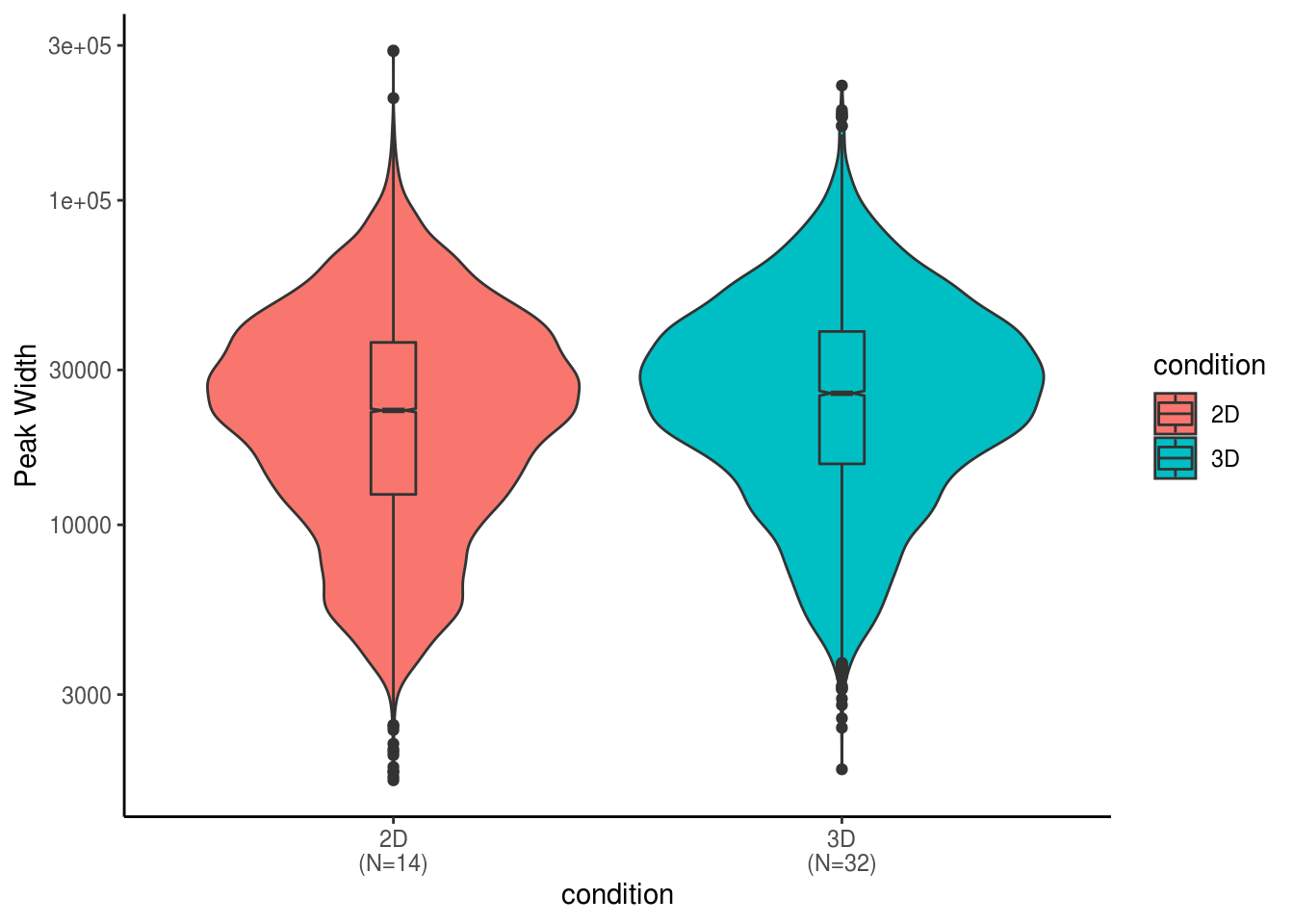
ggplot(peak.summary.df, aes(x=condition, y=numPeak, fill= condition))+ scale_x_discrete(labels=xlabs) +geom_boxplot(width=0.5,position = position_dodge(0.8)) + geom_dotplot(binaxis = "y", stackdir='center', dotsize = 1,position = position_dodge(0.8))+ labs(y="# of Peak") + theme_classic()## Warning: Computation failed in `stat_boxplot()`:
## 'x' must be atomic## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.## Warning: Computation failed in `stat_bindot()`:
## attempt to apply non-function
4.2.3 Super Ehancer rank plot for each samples
Not running it because R can’t render that many plots in one grid
#se.rank.plots <- mapply(get.plot.obj,se.table$se_gene,se.table$all,se.table$sample,SIMPLIFY = FALSE)
#plot_grid( plotlist = se.rank.plots)CCOC.2D.3D.chip.diffbind <- read.csv("data/ChIP-seq/CCOC_2D_3D_chip_diffbind.csv")
CCOC.2D.3D.chip.diffbind$Peaks<- CCOC.2D.3D.chip.diffbind$MACPeak
CCOC.2D.3D.chip.diffbind$PeakCaller <- "narrow"## 3D-HAC2_1 HAC2 3D 1 narrow## 3D-JHOC9_1 JHOC9 3D 1 narrow## 3D_KOC-7C_1 KOC-7C 3D 1 narrow## 3D_KOC-7C_2 KOC-7C 3D 2 narrow## 3D_OV207_1 OV207 3D 1 narrow## 3D_OV207_2 OV207 3D 2 narrow## 3D_OVISE_1 OVISE 3D 1 narrow## 3D_OVISE_2 OVISE 3D 2 narrow## 3D_SMOV2_1 SMOV2 3D 1 narrow## 3D_SMOV2_2 SMOV2 3D 2 narrow## 3D-C5X_1 C5X 3D 1 narrow## 3D-C5X_2 C5X 3D 2 narrow## 3D-HCH1_1 HCH1 3D 1 narrow## 3D-HCH1_2 HCH1 3D 2 narrow## 2D_C5X_1 C5X 2D 1 narrow## 2D_C5X_2 C5X 2D 2 narrow## 2D_ES2_1 ES2 2D 1 narrow## 2D_ES2_2 ES2 2D 2 narrow## 2D_HAC2_1 HAC2 2D 1 narrow## 2D_HAC2_2 HAC2 2D 2 narrow## 2D_HCH1_1 HCH1 2D 1 narrow## 2D_HCH1_2 HCH1 2D 2 narrow## 2D_JHOC5_1 JHOC5 2D 1 narrow## 2D_JHOC5_2 JHOC5 2D 2 narrow## 2D_JHOC9_1 JHOC9 2D 1 narrow## 2D_JHOC9_2 JHOC9 2D 2 narrow## 2D_KOC-7C_1 KOC-7C 2D 1 narrow## 2D_KOC-7C_2 KOC-7C 2D 2 narrow## 2D_OV207_1 OV207 2D 1 narrow## 2D_OV207_2 OV207 2D 2 narrow## 2D_OVAS_1 OVAS 2D 1 narrow## 2D_OVAS_2 OVAS 2D 2 narrow## 2D_OVISE_1 OVISE 2D 1 narrow## 2D_OVISE_2 OVISE 2D 2 narrow## 2D_OVMANA_1 OVMANA 2D 1 narrow## 2D_OVMANA_2 OVMANA 2D 2 narrow## 2D_OVSAYO_1 OVSAYO 2D 1 narrow## 2D_OVSAYO_2 OVSAYO 2D 2 narrow## 2D_OVTOKO_1 OVTOKO 2D 1 narrow## 2D_OVTOKO_2 OVTOKO 2D 2 narrow## 2D_RMGII_1 RMGII 2D 1 narrow## 2D_RMGII_2 RMGII 2D 2 narrow## 2D_SMOV2_1 SMOV2 2D 1 narrow## 2D_SMOV2_2 SMOV2 2D 2 narrow## 2D_TOV21G_1 TOV21G 2D 1 narrow## 2D_TOV21G_2 TOV21G 2D 2 narrow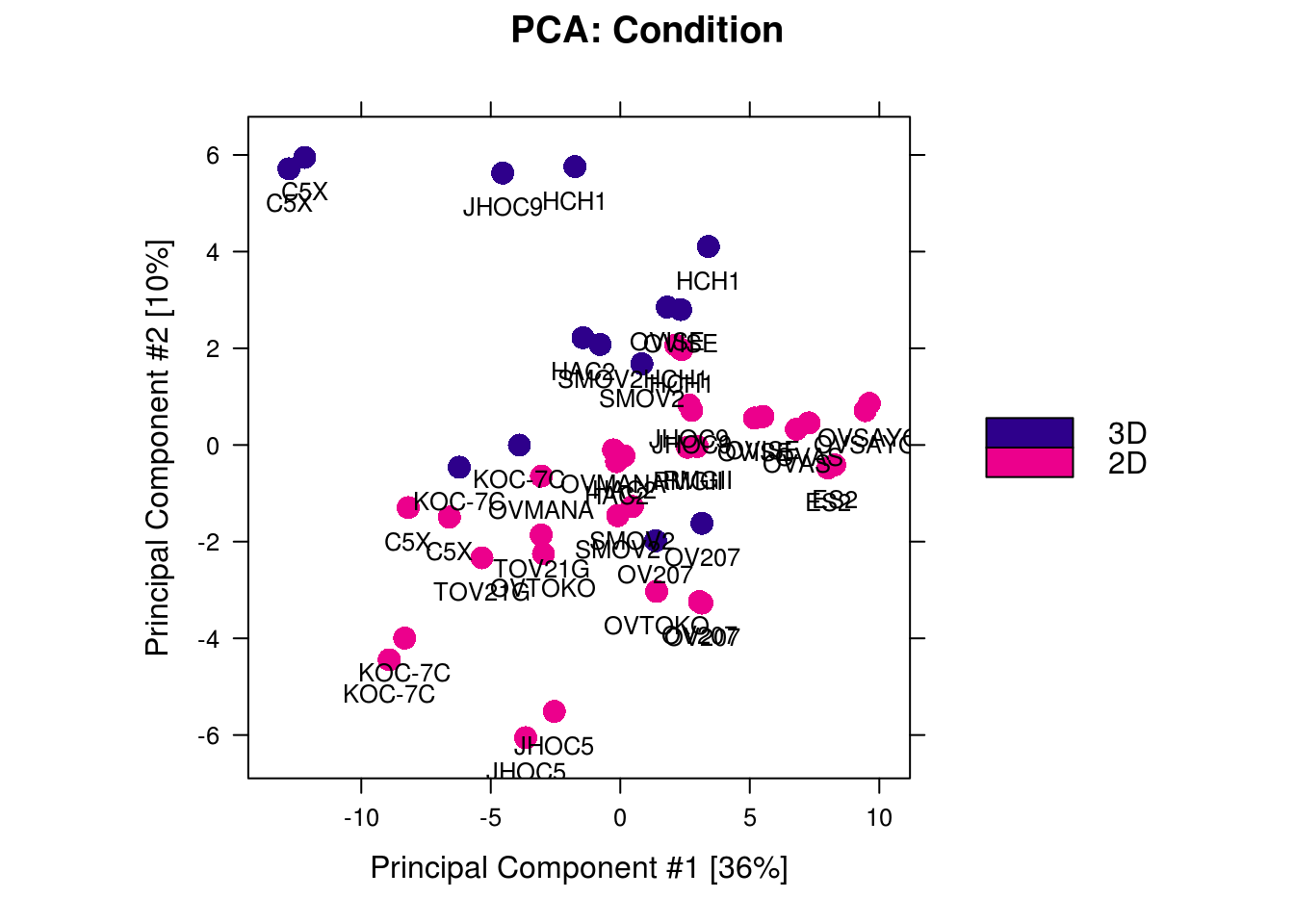
## 1613 genes were dropped because they have exons located on both strands of the same reference sequence or on more than one reference sequence, so cannot be represented by
## a single genomic range.
## Use 'single.strand.genes.only=FALSE' to get all the genes in a GRangesList object, or use suppressMessages() to suppress this message.